| General Information
|
|
| Noonan syndrome, Costello syndrome, cardio-facio-cutaneous syndrome, Noonan syndrome with multiple lentigines (LEOPARD syndrome), Noonan syndrome with loose anagen hair (Mazzanti syndrome), CBL mutation-associated syndrome and related phenotypes, collectively called RASopathies, are all characterized by short stature, heart defects and typical facial anomalies. Other symptoms and severity of clinical expression vary among individual syndromes. Especially in early childhood these conditions may be hard to distinguish from each other.Heterozygous mutations in genes encoding components of the RAS/MAPK pathway have been found to cause Noonan syndrome and related disorders, namely PTPN11, SOS1, RAF1, KRAS, NRAS, HRAS, CBL, BRAF, MEK1, MEK2, SHOC2 and RIT1.
|
|
| The NSEuroNet is a European network on Noonan syndrome and related disorders (RASopathies). One of the main projects the group has been involved in is the development of a genotype-phenotype database for the RASopathies. While the number of mutations and sequence variations identified in RASopathy genes is still increasing, the probability for novel mutations to be reported to the public has become quite low and the significance of rare variations may even remain unclear. Moreover, phenotypic data of published cohorts is hardly comparable due to the lack of standardization, and individual study cohorts do not reach the statistical power for reliable genotype-phenotype correlations.For these reasons the NSEuroNet Consortium has developed a database that contains published germline mutations in the known RASopathy genes (excluding NF1), unpublished mutations observed by the consortium partners and collaborators, as well as polymorphisms and unclassified variations. In addition, standardized clinical datasets on a steadily increasing number of patients with a molecularly proven RASopathy are collected in order to establish genotype-phenotype correlations.The database is freely accessible. It can be browsed for genes, mutations or phenotypes through a user-friendly graphical surface. We introduce this novel resource which will be of great value for scientists as well as for clinical geneticists involved in counseling of patients with these disorders and their families.
|
|
| Our work has been funded through the Federal Ministry of Education and Research in Germany (BMBF) (E-rare (ERA-Net for Research Programs on Rare Diseases) project “NSEuroNet”).
|
|
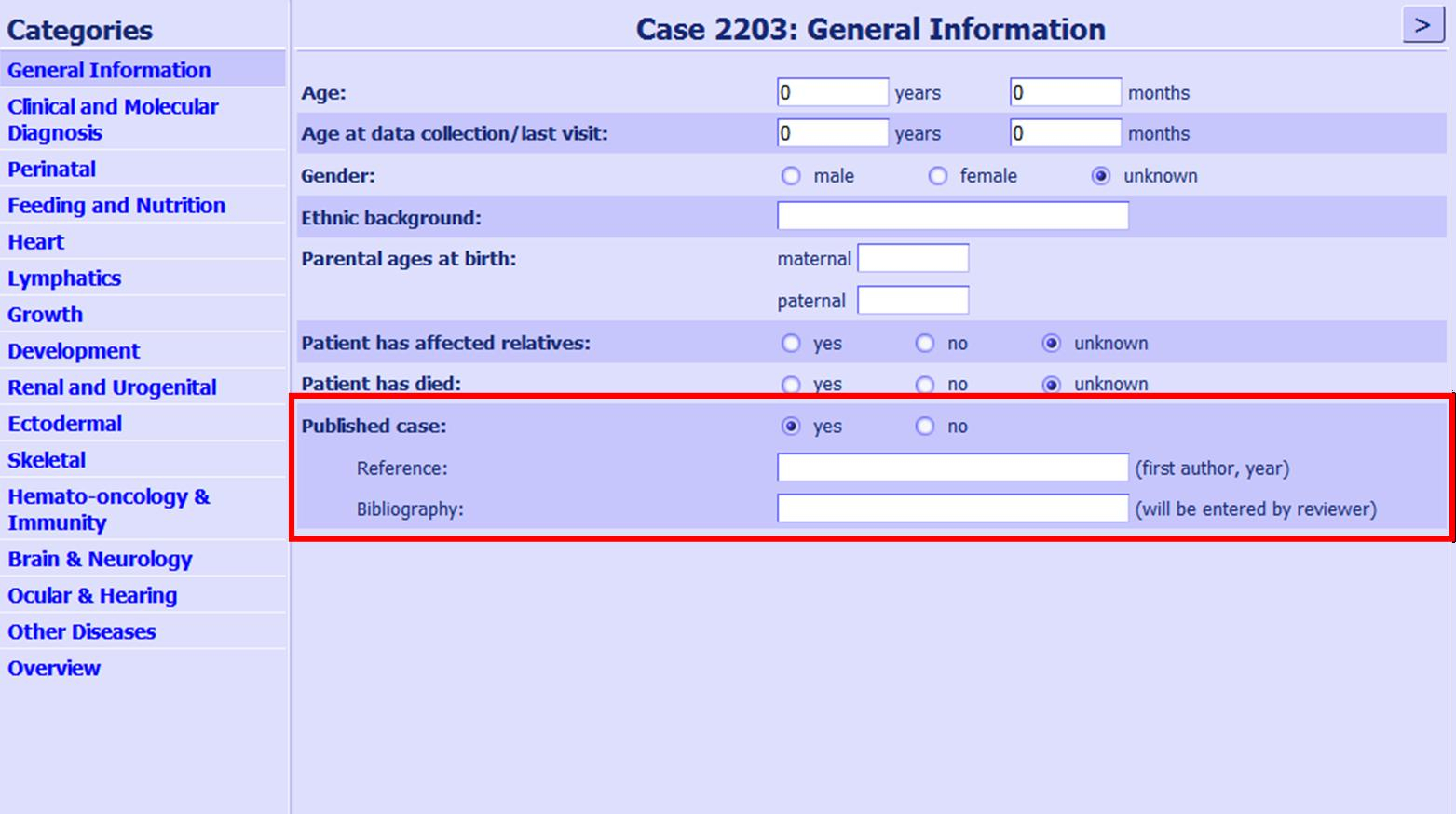
| Genotype and phenotype data can be submitted to the database by registered users and will be added after review by the curators. In principle, any person can register as a submitter, but the inquired data usually requires a medical background. To avoid duplicates in the database we encourage every submitter to take care that the same case is not entered multiple times, e.g. after a follow-up. Every entry receives a unique case number that is created by the system. Only the submitter of an individual dataset is able to link the case number to an individual patient (by recording the case number in the patient’s file). Using this case number, entries can be edited only by the submitter to add follow-up data or to correct inaccurate information. Also, if a patient has been seen by multiple specialists or been part of a study cohort, submitters are encouraged to ensure that he/she has not already been entered by a different person.The database includes published cases that are molecularly proven. Only the basic information (mutation and syndrome) have been added for these patients. If a patient has previously been published, the submitter is asked to indicate this in the respective place on the questionnaire (see figure 1) and we will erase one of our literature patients with the same mutation and syndrome. This will ensure that the quantity of cases remains accurate. We intend to update the literature patients regularly.
|
|
|
Figure 1:Screenshot of the data entry form. The marked position shows, where a publication can be entered, if the patient has been published.
|
| Literature was found through PubMed using general searches for the gene names. The complete reference list can be found by clicking on “Referencelist” on the top of the page.
|
|
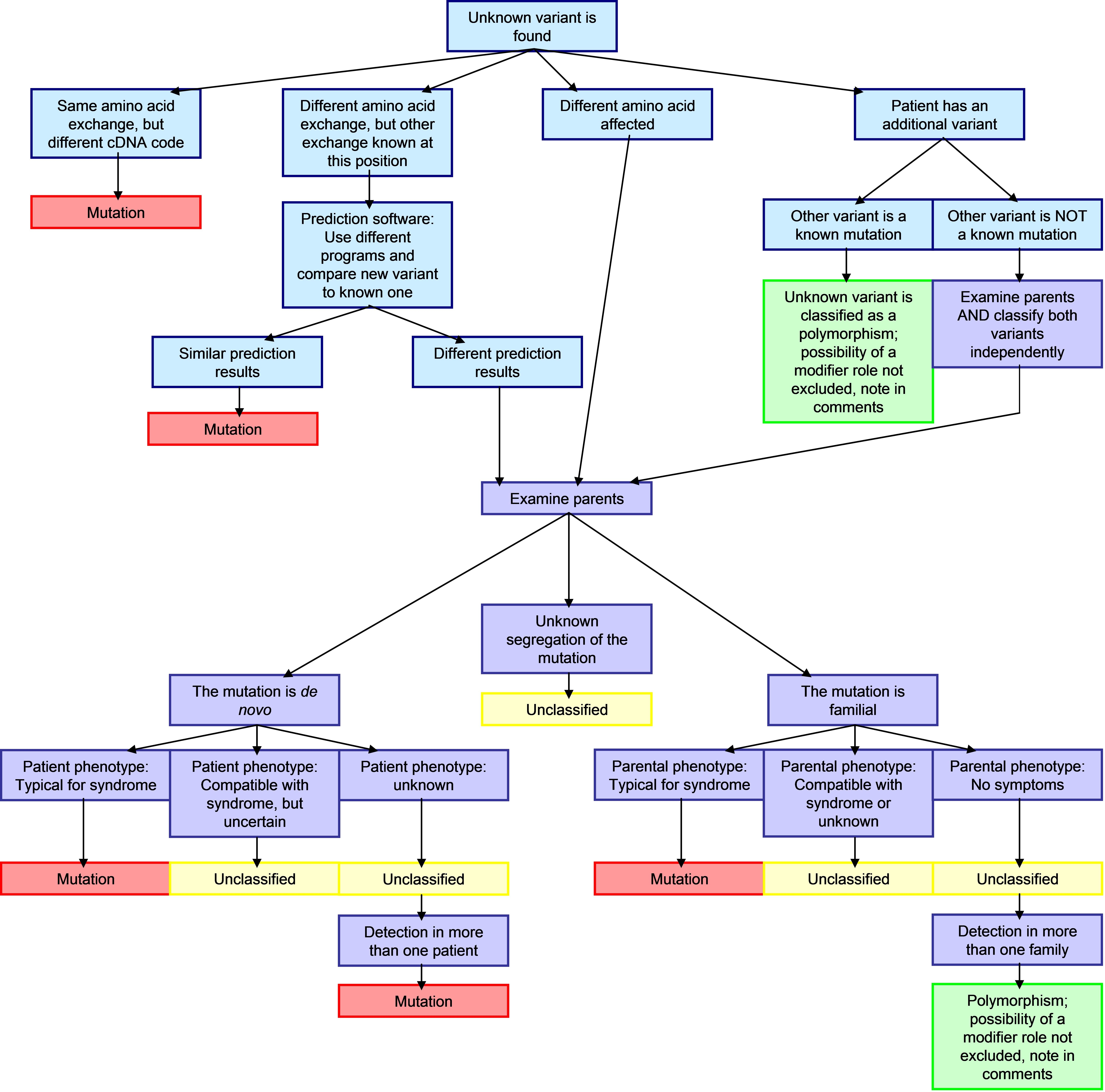
| Another important part of this database is the classification of novel variations. New variations in submitted in this database are analyzed according to the flow chart in figure 2.
|
|
|
Figure 2:Classification flowchart for new variants.
|
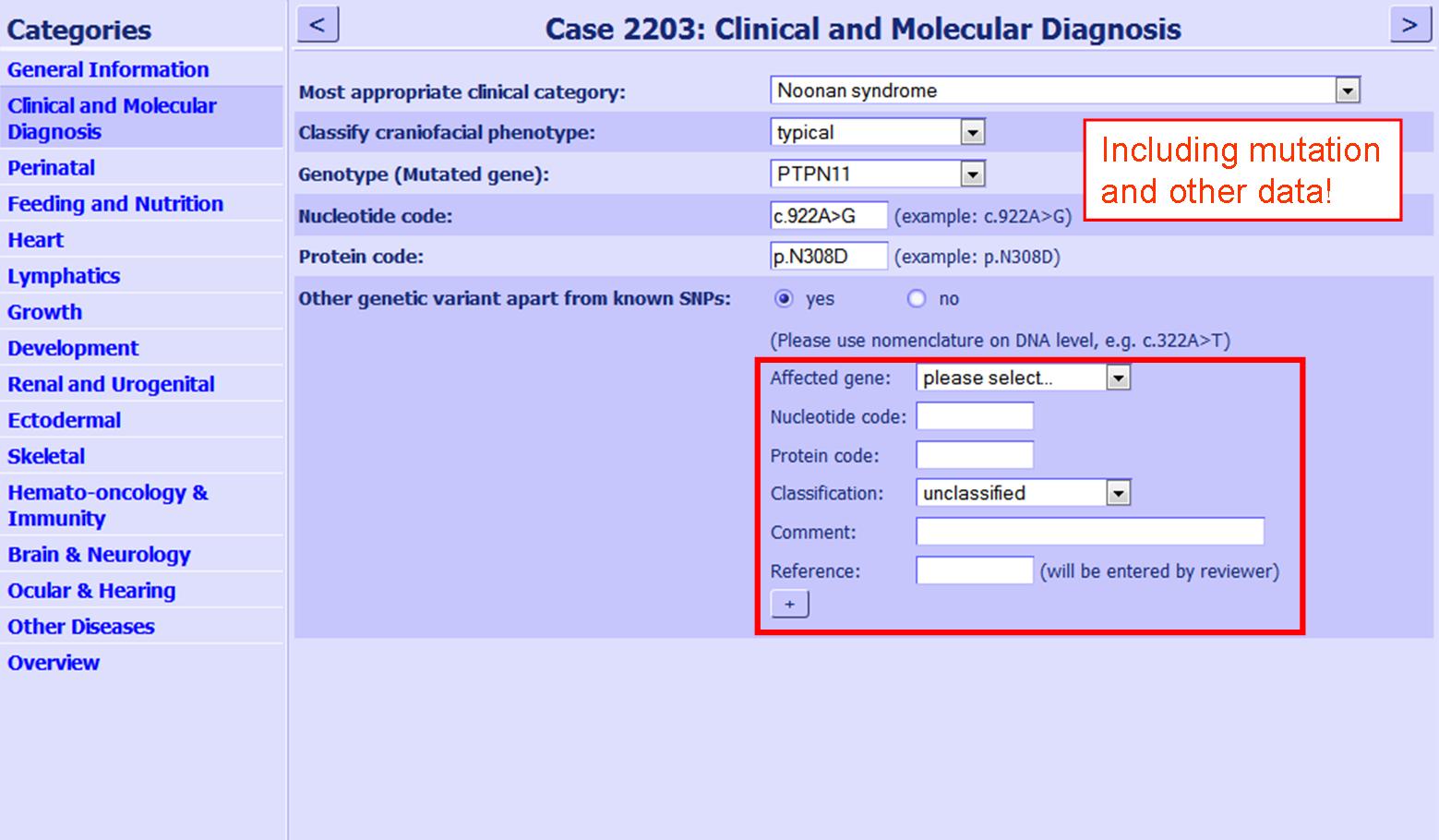
| We encourage the submission of unclassified variations and polymorphisms. If additional variations are found in a patient these can be submitted with the patients data, under the mutation information (see figure 3).
|
|
|
Figure 3:Screenshot variation with additional mutation. The position is shown, where variations should be entered in the questionnaire, if an additional mutation is present.
|
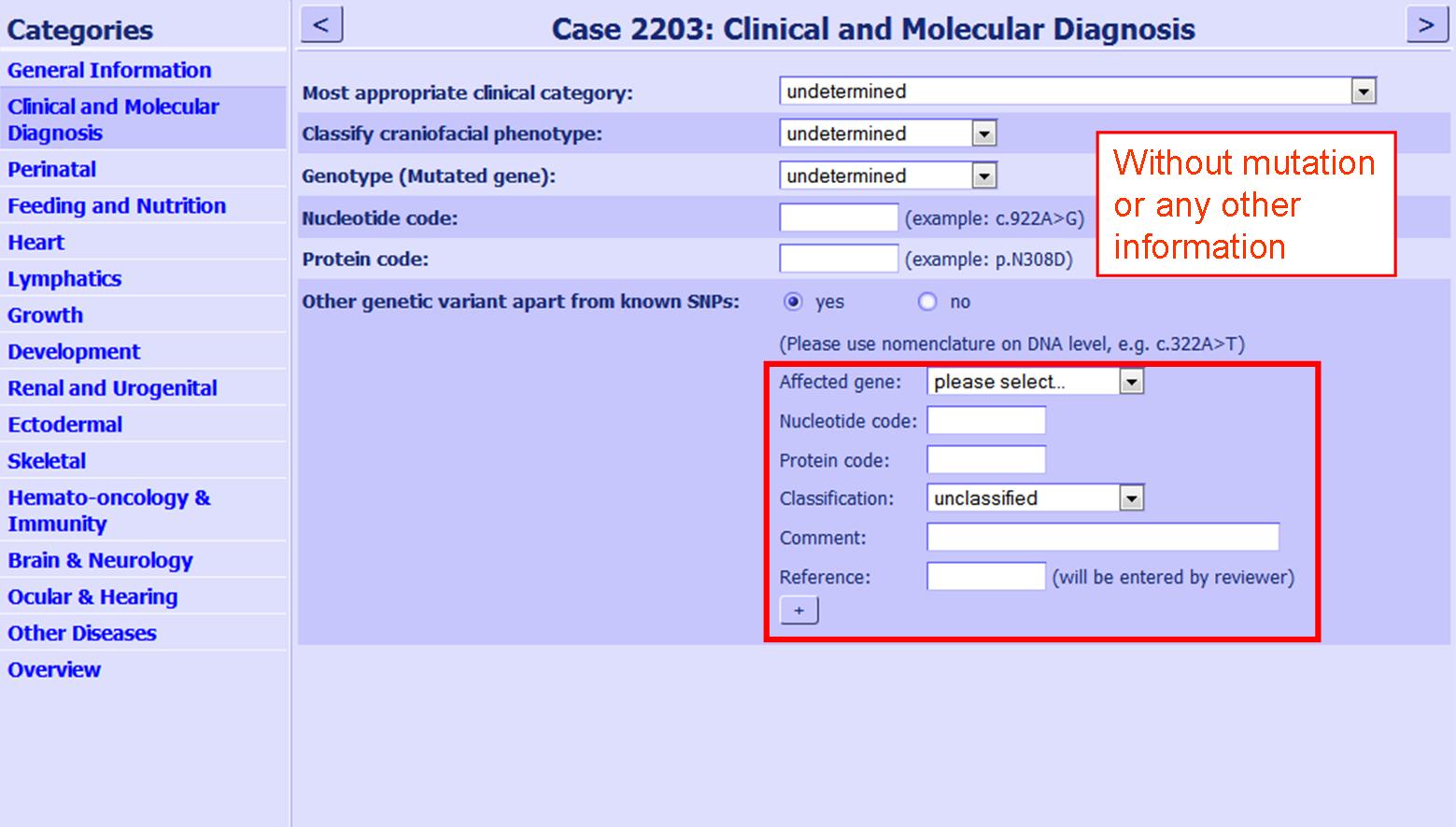
| If a variation is found in an unaffected individual this information can also be entered in a separate data sheet (not with the patients information). A new case can be opened and the only data to be entered is the variation (see figure 4). Please note in the comments that this person is not affected by a RASopathy.
|
|
|
Figure 4:Screenshot variation with out a mutation. The position is shown, where variations should be entered in the questionnaire, if no other mutation has been found for this patient.
|
| In the data output the graphical overview (histogram) only shows mutations. Variations and polymorphisms can be found through the “variations” tab and in the “Details” pop up when clicking on a mutation, amino acid, domain or freely chosen range. The classifications seen here are to the best of our knowledge and may change in time with new results.
|
|
| The NSEuroNet database adheres to standards and recommendations set out by the following groups:- HGNC (HUGO (Human Genome Organisation) Genome Nomenlclature Committee)- HGVS (Human Genome Variation Society)- HVP (Human Variome Project)- LRG (Locus Reference Genomic) |
|
| Please also read our Database Policy and the recommendations about Patient Consent. Information on how to contact the curator can be found in the Impressum / Legal Information contact sections.
|
|




 Welcome,
Home
Referencelist
Caselist
Welcome,
Home
Referencelist
Caselist